|
 |
- Search
| J Environ Anal Health Toxicol > Volume 26(2); 2023 > Article |
|
ABSTRACT
The present study aims to assess the effectiveness of eDNA metabarcoding for the analysis of fish communities in rivers. The findings suggest that the methodology attained an accuracy rate of 80% for species identification and was able to detect between 79.2% and 87.5% of the fish species recorded in the fish monitoring database for three rivers of Anseong, Bokha and Gyeongan during the period 2019 to 2021. Significantly, the eDNA metabarcoding technique enabled the successful detection of comparatively larger fish species, such as Channa argus and Silurus asotus. Furthermore, depending on the river, this method identified 12 to 14 additional species that could not be observed through traditional methodologies. However, it is worth noting that the Mifish primer used amplifies short gene segments, which can pose challenges in identifying species with identical gene sequences. Notwithstanding this limitation, the advantages of eDNA analysis over conventional methods are significant, enabling the identification of a broader range of species within a shorter timeframe, using smaller sample volumes and minimizing risks to both endangered fish and to researchers. As a result, eDNA analysis represents a valuable alternative for assessing biodiversity and for collecting data on fish species that are challenging to analyze.
eDNA (environmental DNA)ļŖö ņāØļ¼╝ņ▓┤Ļ░Ć ļ¼╝, ĒåĀņ¢æ, Ļ│ĄĻĖ░ ļō▒ ņŻ╝ņ£ä ĒÖśĻ▓Įņ£╝ļĪ£ ļ░░ņČ£ĒĢ£ ņ£ĀņĀäļ¼╝ņ¦łņØä ļ¦ÉĒĢśļ®░, ņØ┤ļź╝ ņØ┤ņÜ®ĒĢśļŖö ņ¢┤ļźś ĻĄ░ņ¦æņĪ░ņé¼ (fish community analysis)Ļ░Ć ņäĖĻ│äņĀüņ£╝ļĪ£ ĒÖ£ļ░£ĒĢśĻ▓ī ņŚ░ĻĄ¼ļÉśĻ│Ā ņ׳ļŗż[1]. ņ¢┤ļźśļŖö ļ¼╝ ĒÖśĻ▓ĮņŚÉņä£ ņ▓┤ņĢĪĻ│╝ ņĪ░ņ¦ü ņäĖĒż (tissue)ļź╝ ĒåĄĒĢ┤ņä£ DNAļź╝ ļ░░ņČ£ĒĢśļ®░, ņØ┤ļź╝ ņłśņ¦æĒĢśĻ│Ā ļČäņäØĒĢśļ®┤ ĒĢ┤ļŗ╣ ņ¦ĆņŚŁņØś ĒŖ╣ņĀĢ ņ¢┤ļźśņØś ņĪ┤ņ×¼ ņŚ¼ļČĆ, ĒÆŹļČĆļÅä (abundance) ļ░Å ļŗżņ¢æņä▒ (diversity) ļō▒ņØä ņČöļĪĀĒĢĀ ņłś ņ׳ļŗż[2]. ņ¢┤ļźś ĻĄ░ņ¦æ ļČäņäØņŚÉ eDNAļź╝ ņé¼ņÜ®ĒĢśļ®┤ ņ×Éļ¦Ø (gill net), ņĀäĻĖ░ ļéÜņŗ£ (electrofishing), ņĀĢņ╣śļ¦Ø (fyke nets), ņĪ▒ļīĆ (kick net), Ēł¼ļ¦Ø (casting net) ļō▒ņØä ņØ┤ņÜ®ĒĢśņŚ¼ ņ▒äņ¦æĒĢśĻ│Ā ņĀä ļ¼Ėņ¦ĆņŗØ (taxonomic expertise)ņØ┤ ĒĢäņÜöĒĢ£ ĒśĢĒā£ņĀü ĒŖ╣ņ¦Ģ (morphological characters)ņØä ĒåĄĒĢ┤ ļÅÖņĀĢĒĢśļŖö ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉ ļ╣äĒĢ┤ ņŚ¼ļ¤¼ Ļ░Ćņ¦Ć ņØ┤ņĀÉņØ┤ ņ׳ļŗż. eDNA ņĪ░ņé¼ļ▓ĢņØĆ ļ╣äņ╣©ņŖĄņĀü (non-invasive)ņ£╝ļĪ£ ĒÖśĻ▓ĮĻ│╝ ņ¢┤ļźś Ļ░£ņ▓┤ĻĄ░ņŚÉ ņĄ£ņåīĒĢ£ņØś ņśüĒ¢źņØä ļ»Ėņ╣śĻĖ░ ļĢīļ¼ĖņŚÉ ĒÖśĻ▓Į ļ│┤ņĀä ļ░Å ņāØĒā£ņĪ░ņé¼ļź╝ ņ£äĒĢ£ ņ£ĀņÜ®ĒĢ£ ļÅäĻĄ¼ ņżæņØś ĒĢśļéśņØ┤ļ®░[3], ņĀäĒåĄņĀüņØĖ ļ░®ļ▓Ģņ£╝ļĪ£ Ļ┤Ćņ░░ĒĢśĻĖ░ ņ¢┤ļĀżņÜ┤ ņóģ (rare, elusive, cryptic species)ņØä ĒāÉņ¦ĆĒĢĀ ņłśļÅä ņ׳ļŗż[4]. ļśÉĒĢ£ ņĀäļ¼ĖņĀüņØĖ ļČäļźśĒĢÖņĀü ņ¦ĆņŗØņØ┤ļéś ņŗ£Ļ░äĻ│╝ ņØĖļĀźņØä ļ¦ÄņØ┤ ņåīļ¬©ĒĢśņ¦Ć ņĢŖņĢäļÅä ņÖĖļלņóģ ļśÉļŖö ļ®Ėņóģ ņ£äĻĖ░ ņóģņØś ņĪ┤ņ×¼ļź╝ ĒżĒĢ©ĒĢśņŚ¼ ņ¢┤ļźś ĻĄ░ņ¦æņŚÉ ļīĆĒĢ£ ĒżĻ┤äņĀüņØĖ ĒśäĒÖ®ņØä ņé┤ĒÄ┤ļ│╝ ņłś ņ׳ļŗż[5]. ņØ┤ ļ░®ļ▓ĢņØĆ ĒĢśņ▓£ņŚÉņä£ļČĆĒä░ ņŗ¼ĒĢ┤ ņāØĒā£Ļ│äņŚÉ ņØ┤ļź┤ĻĖ░Ļ╣īņ¦Ć ļŗżņ¢æĒĢ£ ļ¼╝ ĒÖśĻ▓ĮņŚÉņä£ ņä▒Ļ│ĄņĀüņ£╝ļĪ£ ņé¼ņÜ®ļÉśņŚłņ£╝ļ®░ ņ¢┤ļźśņØś ļŗżņ¢æņä▒ ļ░Å ļČäĒżņŚÉ ļīĆĒĢ£ ņÜ░ļ”¼ņØś ņØ┤ĒĢ┤ļź╝ ļäōĒ×É ņłś ņ׳ļŖö ņ×Āņ×¼ļĀźļÅä Ļ░Ćņ¦ĆĻ│Ā ņ׳ļŗż[6]. ĻĄŁļé┤ņŚÉņä£ļŖö ņĄ£ĻĘ╝ ļōżņ¢┤ ļČĆņé░ ņŚ░ņĢł[7], ļÅÖĒĢ┤[8], Ļ▓ĮĻĖ░ļÅä ĒĢ┤ņ¢æņłśņé░ņ×ÉņøÉņŚ░ĻĄ¼ņåī ņāØĒā£ĒĢÖņŖĄĻ┤Ć[9], ĒÖ®ĻĄ¼ņ¦Ćņ▓£[10], 4Ļ░£ ĒĢśņ▓£ (ĻĖłĻ░Ģ, ņ¦Ćņ▓£, ĒÖ®ņ¦Ćņ▓£, ņä¼ņ¦äĻ░Ģ) [11], ļśÉ ļŗżļźĖ 4Ļ░£ ĒĢśņ▓£ (ĒśĢņé░Ļ░Ģ, Ēā£ĒÖöĻ░Ģ, ņä¼ņ¦äĻ░Ģ, ļéÖļÅÖĻ░Ģ) [12] ņŚÉ ļīĆĒĢ£ ņĪ░ņé¼ ņé¼ļĪĆĻ░Ć ņ׳ņ¦Ćļ¦ī ņĢäņ¦üņØĆ ĒÖ£ļ░£ĒĢśĻ▓ī ņłśĒ¢ēļÉśĻ│Ā ņ׳ņ¦Ć ņĢŖļŗż.
ļ│Ė ņŚ░ĻĄ¼ļŖö Ļ▓ĮĻĖ░ļÅä ļé┤ 3Ļ░£ ĒĢśņ▓£ņØä ļīĆņāüņ£╝ļĪ£ ņ¢┤ļźś ĻĄ░ņ¦æ ļČäņäØņŚÉ eDNAņØś ĒÖ£ņÜ® Ļ░ĆļŖźņä▒ņØä Ļ▓ĆĒåĀĒĢśņśĆļŗż. ņ¢┤ļźśļŖö ĒĢśņ▓£ņŚÉņä£ļŖö ņāØĒā£ Ļ▒┤Ļ░Ģņä▒ ĒÅēĻ░ĆņØś ņ¦ĆĒæ£ļĪ£, ĒśĖņåīņŚÉņä£ļŖö ņāØļ¼╝ ļŗżņ¢æņä▒ ņ¦ĆĒæ£ļĪ£ņä£ ĒÖ£ņÜ®ļÉśĻ│Ā ņ׳ņ£╝ļéś ņ¦Ćņ×Éņ▓┤ ņŚ░ĻĄ¼ĻĖ░Ļ┤Ć ļō▒ņŚÉņä£ ļŖö ņĀäļ¼ĖņØĖļĀźņØś ļČĆņĪ▒ņ£╝ļĪ£ ņØ┤ļ¤¼ĒĢ£ ņĪ░ņé¼ļź╝ ņĀ£ļīĆļĪ£ ņłśĒ¢ēĒĢśņ¦Ć ļ¬╗ĒĢśļŖö ņŗżņĀĢņØ┤ļŗż. ļö░ļØ╝ņä£ ņØ┤ļ▓ł ņŚ░ĻĄ¼ļŖö ņ¢┤ļźś ņĪ░ņé¼ņŚÉ eDNA ļ®öĒāĆļ░öņĮöļö®ņØś ņ£ĀņÜ®ņä▒ņØä ĒÅēĻ░ĆĒĢśņŚ¼ Ē¢źĒøä ļ¼╝ĒÖśĻ▓ĮņāØĒā£ ņĪ░ņé¼ņŚÉ ĒÖ£ņÜ®ĒĢśĻ│Āņ×É ņłśĒ¢ēļÉśņŚłļŗż.
ņĪ░ņé¼ ļīĆņāüņØĆ Ļ▓ĮĻĖ░ļÅä ĒĢ┤ņ¢æņłśņé░ņ×ÉņøÉņŚ░ĻĄ¼ņåī ņāØĒā£ĒĢÖņŖĄĻ┤Ć (Ļ▓ĮĻĖ░ļÅä ņ¢æĒÅēĻĄ░ ņÜ®ļ¼Ėļ®┤) ļ░Å ĒÖśĻ▓ĮļČĆ ņāØļ¼╝ņĖĪņĀĢļ¦Ø ņ¦ĆņĀÉņØĖ ņĢłņä▒ņ▓£1 (ņĢłņä▒ņŗ£ ĒśäņłśļÅÖ, ņĢłņä▒ĻĄÉ), ļ│ĄĒĢśņ▓£1-1 (ņØ┤ņ▓£ņŗ£ Ļ░łņé░ļÅÖ, ļ│ĄĒĢś2ĻĄÉ), Ļ▓ĮņĢłņ▓£5 (Ļ┤æņŻ╝ņŗ£ ņ┤łņøöņØŹ ļ¼┤Ļ░æļ”¼, ņä£ĒĢśĻĄÉ)ļź╝ ņäĀņĀĢĒĢśņśĆĻ│Ā (Fig. 1), ĒĢśņ▓£ņłś ņŗ£ļŻīļŖö ņāØļ¼╝ņĖĪņĀĢļ¦Ø (2019~2021) Ļ░ĆņØäņ▓Ā ņ×ÉļŻīņÖĆ ļ╣äĻĄÉĒĢśĻ│Āņ×É 9~10ņøöņŚÉ ņ▒äņĘ©ĒĢśņŚ¼ ļČäņäØĒĢśņśĆļŗż.
ļ│Ė ņŚ░ĻĄ¼ņŚÉņä£ļŖö eDNA ļČäņäØļ▓ĢņØä ĒĢśņ▓£ņŚÉ ņĀüņÜ®ĒĢśĻĖ░ ņĀäņŚÉ ņŗżĒŚśņĪ░Ļ▒┤ ļ░Å ņĀüņÜ® Ļ░ĆļŖźņä▒ņØä ĒÖĢņØĖĒĢśĻĖ░ ņ£äĒĢ┤ Ļ▓ĮĻĖ░ļÅä ĒĢ┤ņ¢æņłśņé░ņ×ÉņøÉņŚ░ĻĄ¼ņåīņŚÉņä£ ņÜ┤ņśüĒĢśļŖö ļ»╝ļ¼╝Ļ│ĀĻĖ░ ņāØĒā£ĒĢÖņŖĄĻ┤ĆņŚÉņä£ ņ×äņØśļĪ£ ņäĀņĀĢĒĢ£ ĒĢśļéśņØś ņóģņØ┤ ņä£ņŗØĒĢśļŖö ņłśņĪ░ 20Ļ││ņŚÉņä£ ņŗ£ļŻī 200 mLļź╝ Ļ░üĻ░ü ņ▒äņĘ©ĒĢśņśĆļŗż. ņŗżĒŚśņŗż ņØ┤ļÅÖ Ēøä ņ▒äņĘ©ĒĢ£ 20Ļ░£ ņŗ£ļŻīļŖö Ļ░ÖņØĆ ņ¢æņ£╝ļĪ£ Ēś╝ĒĢ®ĒĢ£ Ēøä 0.45-╬╝m Micro Funnel filter (Pall Life Science, Puerto Rico)ļĪ£ ņŚ¼Ļ│╝ĒĢśĻ│Ā DNeasy PowerWater kit (Qiagen, Hiden, Germany) ļź╝ ņØ┤ņÜ®ĒĢ┤ eDNAļź╝ ņČöņČ£ĒĢśņśĆļŗż. ņČöņČ£ĒĢ£ eDNAņØś ņ¢┤ļźś ļ®öĒāĆļ░öņĮöļö® (metabarcoding)ņØĆ ļ»ĖĒåĀņĮśļō£ļ”¼ņĢä 12S rRNA ņ£ĀņĀäņ×ÉņØś ļ│ĆņØ┤Ļ░Ć ļ¦ÄņØĆ ņśüņŚŁ (hypervariable region)ņØä ļīĆņāüņ£╝ļĪ£ ĒĢśļŖö Mifish U-FņÖĆ Mifish U-R primer [14]ņÖĆ Illumina MiSeq platformļź╝ ņØ┤ņÜ®ĒĢśņŚ¼ ļČäņäØņØä ņłśĒ¢ēĒĢśņśĆļŗż (Macrogen, Seoul).
ĒĢśņ▓£ ņŗ£ļŻīļŖö ļ®ĖĻĘĀĒĢ£ ņÜ®ĻĖ░ļź╝ ņØ┤ņÜ®ĒĢ┤ ļŗżļ”¼ ņ£äņŚÉņä£ ņ▒äņĘ©ĒĢśĻ│Ā 50-mL ņØ╝ĒÜīņÜ® ņŻ╝ņé¼ĻĖ░ņÖĆ 0.45-╬╝m Sterivex filter (Merck Millipore, Darmstadt, Germany)ļź╝ ņØ┤ņÜ®ĒĢ┤ ĒśäņןņŚÉņä£ ~ 500 mL ņĀĢļÅäļź╝ ņŚ¼Ļ│╝ĒĢ£ Ēøä ļō£ļØ╝ņØ┤ņĢäņØ┤ņŖżļź╝ ņØ┤ņÜ®ĒĢ┤ ņĀĆņś© ļ│┤Ļ┤Ć ņÜ┤ļ░śĒĢśņŚ¼ ļ®öĒāĆļ░öņĮöĒīģ ļČäņäØņØä ņłśĒ¢ēĒĢśņśĆļŗż (NICEM. Seoul). eDNA ņČöņČ£ņØĆ DNeasy Blood & Tissue kit (Qiagen, Hiden, Germany)ļź╝ ņØ┤ņÜ®ĒĢśņśĆņ£╝ļ®░ ļéśļ©Ėņ¦ĆļŖö ņĢ×ņä£ņØś Ļ│╝ņĀĢĻ│╝ Ļ░Öļŗż. ļ®öĒāĆļ░öņĮöļö®ņ£╝ļĪ£ļČĆĒä░ ņ¢╗ņ¢┤ņ¦ä FASTA ĒīīņØ╝ņØĆ Mifish pipeline [15]ņØä ņØ┤ņÜ®ĒĢśņŚ¼ primer ņä£ņŚ┤Ļ│╝ ĒÆłņ¦łņØ┤ ļé«ņØĆ ņä£ņŚ┤ ņĀ£Ļ▒░ļź╝ ĒåĄĒĢ┤ 97% ņØ┤ņāüņØś ņ£Āņé¼ņä▒ (similarity)ņØä Ļ░Ćņ¦ä ņä£ņŚ┤ļōżņØä MOTUs (molecular operational taxo-nomic units)ļĪ£ ņäĀņĀĢĒĢśĻ│Ā BlastnņØä ĒåĄĒĢ┤ NCBI (national center for biotechnology information)ņØś ļŹ░ņØ┤Ēä░ļ▓ĀņØ┤ņŖż (GenBank)ņÖĆ ļ╣äĻĄÉĒĢśņŚ¼ Ļ░ÖņØĆ ņ£Āņé¼ļÅäļź╝ Ļ░Ćņ¦ä 2Ļ░£ ņØ┤ņāüņØ┤ ņ¢╗ņ¢┤ņ¦Ćļ®┤ ĻĄŁļé┤ ņä£ņŗØ ņ¢┤ņóģņ£╝ļĪ£ ļÅÖņĀĢĒĢśņśĆļŗż. eDNAņÖĆ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉ ņØśĒĢ£ ņ¢┤ļźś ĻĄ░ņ¦æ ņĪ░ņé¼ Ļ▓░Ļ│╝ņÖĆ ļ╣äĻĄÉĒĢśĻĖ░ ņ£äĒĢ┤ ĒÖśĻ▓ĮļČĆ ļ¼╝ĒÖśĻ▓ĮņĀĢļ│┤ņŗ£ņŖżĒģ£ņØś 2019~2021ļģä 2ņ░© ņĪ░ņé¼ (Ļ░ĆņØäņ▓Ā)ņŚÉ ĒĢ£ļ▓ł ņØ┤ņāü ņ▒äņ¦æļÉ£ ņ¢┤ņóģĻ│╝ ņØ┤ļ▓ł ņŚ░ĻĄ¼ņŚÉņä£ eDNAņŚÉņä£ ļÅÖņĀĢļÉ£ ņ¢┤ņóģņØä Ēæ£ņŚÉ ĒĢ©Ļ╗ś ļéśĒāĆļé┤ņŚłļŗż. ļśÉĒĢ£ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉ ņØśĒĢ┤ ņ▒äņ¦æļÉ£ Ļ░ü ņ¢┤ļźś ņóģņØś Ļ░£ņ▓┤ ņłśņÖĆ eDNA ļ®öĒāĆļ░öņĮöļö®ņØä ĒåĄĒĢ┤ ņ¢╗ņ¢┤ņ¦ä readņØś Ļ░£ņłśļź╝ ņØ┤ņÜ®ĒĢ┤ ļŗżņ¢æņä▒ ņ¦Ćņłś (H`, ShannonŌĆÖs diversity index)ņÖĆ ĻĘĀļō▒ļÅä ņ¦Ćņłś (E, PielouŌĆÖs evenness index)ļź╝ ļŗżņØīĻ│╝ Ļ░ÖņØ┤ Ļ│äņé░ĒĢśņŚ¼ ļ╣äĻĄÉĒĢśņśĆļŗż.
ņŚ¼ĻĖ░ņä£, Pi = ni/N (ni = ĒĢ£ ņóģņØś Ļ░£ņ▓┤ņłś, N = ļ¬©ļōĀ ņóģņØś ņ┤Ø Ļ░£ņ▓┤ņłś)
ņŚ¼ĻĖ░ņä£, H` = ļŗżņ¢æņä▒ ņ¦Ćņłś, Hmax = ln S (S = ļ░£Ļ▓¼ļÉ£ ņóģ ņłś)
ļ│Ė ņŚ░ĻĄ¼ņŚÉņä£ļŖö Mifish primerļź╝ ņØ┤ņÜ®ĒĢśņŚ¼ Ļ▓ĮĻĖ░ļÅä ĒĢ┤ņ¢æņłśņé░ņ×ÉņøÉņŚ░ĻĄ¼ņåī ņāØĒā£ĒĢÖņŖĄĻ┤ĆņŚÉņä£ ņ×äņØśļĪ£ ņäĀņĀĢĒĢ£ 20Ļ░£ ļīĆņāü ņ¢┤ņóģ ņżæņŚÉņä£ 16Ļ░£ ņóģņØ┤ ĒÖĢņØĖļÉśņ¢┤ 80%Ļ░Ć Ļ▓ĆņČ£ Ļ░ĆļŖźĒĢśņśĆļŗż (Table 1). ņØ┤ļ¤¼ĒĢ£ Ļ▓ĆņČ£ņ£©ņØĆ ņĢ×ņä£ ņ¦äĒ¢ēļÉ£ ņŚ░ĻĄ¼[9]ņØś 90% ļ│┤ļŗż ļé«ņĢśņ£╝ļ®░ ļ¬ć Ļ░Ćņ¦Ć ņøÉņØĖņØä Ļ│ĀļĀżĒĢĀ ņłś ņ׳ņ£╝ļéś[16-18] eDNAļź╝ ņČöņČ£ĒĢśĻĖ░ ņ£äĒĢ┤ ņé¼ņÜ®ļÉ£ kitņØś ņ░©ņØ┤ ļĢīļ¼Ėņ£╝ļĪ£ ņČöņĀĢļÉ£ļŗż. ņØ┤ņĀä ņŚ░ĻĄ¼ņŚÉņä£ ņ¢┤ļ¢ż ņóģļźśņØś kitĻ░Ć ņé¼ņÜ®ļÉśņŚłļŖöņ¦Ć ņĀ£ņŗ£ļÉśņ¦Ć ņĢŖņĢśņ¦Ćļ¦ī ņØ┤ļ▓ł ņŚ░ĻĄ¼ņŚÉņä£ļŖö ĒĢśņ▓£ ņŗ£ļŻī eDNA ņČöņČ£ņŚÉ DNeasy PowerWater kit (PW)ļź╝ ņé¼ņÜ®ĒĢ£ Ļ▓ĮņÜ░ 3ļ▓ł ņŚ░ņåŹ ļ®öĒāĆļ░öņĮöļö® ļČäņäØņØ┤ ņĀ£ļīĆļĪ£ ņ¦äĒ¢ēļÉśņ¦Ć ņĢŖņĢśņ¦Ćļ¦ī DNeasy Blood & Tissue kit (BT)ļź╝ ņØ┤ņÜ®ĒĢ£ Ļ▓ĮņÜ░ņŚÉļŖö ņĀüņĀłĒĢ£ Ļ▓░Ļ│╝ļź╝ ņ¢╗ņØä ņłś ņ׳ņŚłļŗż. ļśÉĒĢ£ ņāØĒā£ĒĢÖņŖĄĻ┤Ć ņŗ£ļŻīņŚÉņä£ Ļ▓ĆņČ£ĒĢśņ¦Ć ļ¬╗ĒĢ£ Tanakia koreensis (ņ╣╝ļé®ņ×ÉļŻ©) [9], Coreoperca herzi (Ļ║Įņ¦Ć) ļŖö ļŗżļźĖ ņŚ░ĻĄ¼ņŚÉņä£ļŖö Mifish primerņŚÉ ņØśĒĢ┤ Ļ▓ĆņČ£ļÉśļŖö Ļ▓āņ£╝ļĪ£ ļéśĒāĆļé¼Ļ│Ā [16] Koreocobitis rotundicaudata (ņāłņĮöļ»ĖĻŠĖļ”¼)ļŖö Ļ▓ĮņĢłņ▓£ņŚÉņä£ Rhodeus notatus (ļ¢Īļé®ņżäĻ░▒ņØ┤)ļŖö ņĢłņä▒ņ▓£Ļ│╝ ļ│ĄĒĢśņ▓£ņŚÉņä£ Ļ░üĻ░ü Ļ▓ĆņČ£ļÉśņŚłļŗż. eDNAļź╝ ņØ┤ņÜ®ĒĢ£ ņ¢┤ļźś ĻĄ░ņ¦æņŚ░ĻĄ¼ņŚÉļŖö BTņÖĆ PW kitĻ░Ć ņóŗņØĆ ĒÆłņ¦łņØś eDNAļź╝ ņ¢╗ņØä ņłś ņ׳ņ¢┤ņä£ ļ¦ÄņØ┤ ņé¼ņÜ®ļÉśļ®░ [19] DNA extraction kitĻ░Ć eDNA ļ®öĒāĆļ░öņĮöļö®ņŚÉ ļ»Ėņ╣śļŖö ņśüĒ¢źņŚÉ Ļ┤ĆĒĢ£ ņŚ░ĻĄ¼ņŚÉņä£ļÅä BT kitņÖĆ phenol/chloroform extraction ļ░®ļ▓ĢņØä ĒĢ©Ļ╗ś ņĀüņÜ®ĒĢśņŚ¼ Ļ░Ćņן ņóŗņØĆ Ļ▓░Ļ│╝ļź╝ ņ¢╗ņŚłņ£╝ļ®░[20], ņØ┤ļ▓ł ņŚ░ĻĄ¼ņŚÉņä£ļÅä ņĪ░Ļ▒┤Ļ│╝ ļ¬®ņĀüņŚÉ ļ¦×Ļ▓ī DNA extraction kitļź╝ ņäĀĒāØĒĢ┤ņĢ╝ ĒĢ£ļŗżļŖö Ļ▓āņØä ļ│┤ņŚ¼ņŻ╝ņŚłļŗż[18].
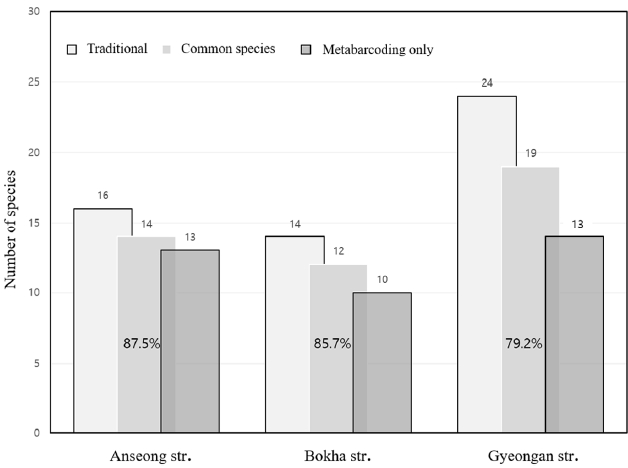
eDNAļź╝ ņØ┤ņÜ®ĒĢ£ ļ│Ė ņŚ░ĻĄ¼ņÖĆ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņØś ĒÖśĻ▓ĮļČĆ ņāØļ¼╝ņĖĪņĀĢļ¦Ø (2019~2021ļģä)ņØś 2ņ░©(Ļ░ĆņØäņ▓Ā) Ļ▓░Ļ│╝ņÖĆ ļ╣äĻĄÉĒĢśņśĆĻ│Ā 3ļģä ņżæņŚÉņä£ ĒĢ£ļ▓łņØ┤ļØ╝ļÅä Ļ┤Ćņ░░ļÉ£ ņ¢┤ļźśļź╝ ļīĆņāüņ£╝ļĪ£ ĒĢśņśĆļŗż (Table 2). Ēæ£ņŚÉļŖö NCBI ņ£ĀņĀäņ×É ņä£ņŚ┤ļČäņäØņŚÉņä£ Ļ░ÖņØĆ ņ£Āņé¼ļÅä(%)ņØś ĻĄŁļé┤ ņä£ņŗØ ņ¢┤ņóģņØ┤ 2Ļ░£ ņØ┤ņāüņØ┤ ņØ╝ņ╣śĒĢśļ®┤ ļ¬©ļæÉ Ēæ£ņŗ£ĒĢśņśĆļŗż. ņśłļź╝ ļōżņ¢┤ Acheilognathus rhombeus/A. gracilis (ļé®ņ¦Ćļ”¼/Ļ░Ćņŗ£ļé®ņ¦Ćļ”¼), Hemibarbus maculatus/Hemi-barbus labeo (ņĢīļØĮļłäņ╣ś/ļłäņ╣ś), Tridentiger obscurus/Tridentiger brevispinis (Ļ▓ĆņĀĢļ¦Øļææ/ļ»╝ļ¼╝Ļ▓ĆņĀĢļ¦Øļææ), Acheilo-gnathus macropterus/A. chankaensis (Ēü░ļé®ņ¦Ćļ”¼/Ļ░Ćņŗ£ļé®ņ¦Ćļ”¼), Oryzias sinensis/Oryzias latipes (ļīĆļźÖņåĪņé¼ļ”¼/ņåĪņé¼ļ”¼)ļō▒ņØĆ Mifish primerņŚÉ ņØśĒĢ┤ ņ”ØĒÅŁļÉ£ ļ»ĖĒåĀņĮśļō£ļ”¼ņĢä 12SrRNA ņ£ĀņĀäņ×É ņŚ╝ĻĖ░ņä£ņŚ┤ļĪ£ļŖö ĻĄ¼ļČäĒĢĀ ņłś ņŚåĻĖ░ ļĢīļ¼ĖņŚÉ ņåŹ (genus) ņłśņżĆņŚÉņä£ļ¦ī ļÅÖņĀĢņØ┤ Ļ░ĆļŖźĒĢśņśĆļŗż. ņØ┤ Ļ░ĆņÜ┤ļŹ░ņŚÉņä£ Ļ▓ĆņĀĢļ¦Øļææ/ļ»╝ļ¼╝Ļ▓ĆņĀĢļ¦ØļææņØĆ ņØ╝ļČĆ ņä▒ņ▓┤ļź╝ ņĀ£ņÖĖĒĢśĻ│ĀļŖö ļČäļźśĒĢÖ ņĀüļÅÖņĀĢ ĒśĢņ¦ł (taxonomic keys)ņØ┤ ļ¬ģĒÖĢĒĢśĻ▓ī ļéśĒāĆļéśņ¦Ć ņĢŖņĢä ņ£ĪņĢłņ£╝ļĪ£ļÅä ļČäļźśĒĢśĻĖ░ ļ¦żņÜ░ ņ¢┤ļĀżņÜ┤ ĻĘ╝ņŚ░ņóģņØ┤ļŗż. 2019 ~ 2021ļģä ņāØļ¼╝ņĖĪņĀĢļ¦ØņŚÉ ļö░ļź┤ļ®┤ 3Ļ░£ ĒĢśņ▓£ņŚÉņä£ņØś ņČ£Ēśäņóģ ņłśļŖö ņĢłņä▒ņ▓£ 12 ~ 14ņóģ, ļ│ĄĒĢśņ▓£ 8 ~ 11ņóģ, Ļ▓ĮņĢłņ▓£ 12 ~ 16ņóģņ£╝ļĪ£ ņØ┤ļ▓ł ņŚ░ĻĄ¼ņØś eDNA ļČäņäØ ļ░®ļ▓ĢņØä ĒåĄĒĢ┤ Ļ▓ĆņČ£ļÉ£ ņ¢┤ņóģņØś 1/2 ņłśņżĆņØ┤ņŚłļŗż. ņĢłņä▒ņ▓£ņŚÉņä£ļŖö ņĀäĒåĄņĀüņØĖ ļ░®ļ▓Ģņ£╝ļĪ£ 3Ļ│╝ (family) 16ņóģ (species) Ļ┤Ćņ░░ļÉśņŚłņ£╝ļ®░ eDNAņŚÉņä£ļŖö ņØ┤ ņżæņŚÉņä£ 14ņóģņØ┤ ĒÖĢņØĖļÉśņŚłĻ│Ā (14/16, 87.5%), 13ņóģņØ┤ ņāłļĪŁĻ▓ī Ļ▓ĆņČ£ļÉśņŚłļŗż. ļ│ĄĒĢśņ▓£ņØś Ļ▓ĮņÜ░, ņāØļ¼╝ņĖĪņĀĢļ¦ØņŚÉņä£ 4Ļ│╝ 14ņóģņØ┤ ņČ£ĒśäĒĢśņśĆņ£╝ļ®░ 12ņóģņØ┤ eDNA ļČäņäØņŚÉņä£ Ļ│ĄĒåĄņĀüņ£╝ļĪ£ ļÅÖņĀĢļÉśņŚłĻ│Ā (12/14, 85.7%) 10ņóģņØś ņāłļĪ£ņÜ┤ ņ¢┤ļźś ņóģņØ┤ Ļ▓ĆņČ£ļÉśņŚłļŗż. Ļ▓ĮņĢłņ▓£ņŚÉņä£ļŖö ņĀäĒåĄņĀüņØĖ ļ░®ļ▓Ģņ£╝ļĪ£ 2Ļ│╝ 24ņóģņØ┤ eDNA ļČäņäØņ£╝ļĪ£ļŖö 11Ļ│╝ 32ņóģņØ┤ ļÅÖņĀĢļÉśņŚłĻ│Ā ņØ┤ ņżæņŚÉņä£ 19ņóģ (19/24, 79.2%)ņØĆ Ļ│ĄĒåĄņĀüņ£╝ļĪ£ ĒÖĢņØĖļÉśņŚłņ£╝ļ®░ 13ņóģņØ┤ ņāłļĪŁĻ▓ī Ļ▓ĆņČ£ļÉśņ¢┤ ĻĖ░ņĪ┤ ņŚ░ĻĄ¼[10]ņØś 35~45%ņŚÉ ļ╣äĒĢ┤ņä£ ņøöļō▒Ē׳ ļåÆņĢśļŗż. ļö░ļØ╝ņä£ ļæÉ ļ░®ļ▓ĢņØä ĒåĄĒĢ┤ ņĢłņä▒ņ▓£ 29ņóģ, ļ│ĄĒĢśņ▓£ 25ņóģ, Ļ▓ĮņĢłņ▓£ 37ņóģņ£╝ļĪ£ 3Ļ░£ ĒĢśņ▓£ņŚÉņä£ ņ┤Ø 47ņóģņØś ņ¢┤ļźśĻ░Ć ĒÖĢņØĖļÉśņŚłļŗż (Fig. 2). ņØ┤ Ļ░ĆņÜ┤ļŹ░ņŚÉņä£ Acheilognathus rhombeus (ļé®ņ¦Ćļ”¼), Carassius auratus (ļČĢņ¢┤), Cyprinus carpio (ņ×ēņ¢┤), Micro-physogobio yaluensis (ļÅīļ¦łņ×É), Odontobutis interrupta (ņ¢╝ļĪØļÅÖņé¼ļ”¼), Pseudogobio esocinus (ļ¬©ļלļ¼┤ņ¦Ć), Pseudoras-bora parva (ņ░ĖļČĢņ¢┤), Rhinogobius sp. (ļ░Ćņ¢┤), Zacco platypus (Ēö╝ļØ╝ļ»Ė)ņÖĆ Ļ░ÖņØĆ 7ņóģņØĆ 3Ļ░£ ĒĢśņ▓£ņŚÉņä£ ļæÉ ļ░®ļ▓Ģ ļ¬©ļæÉņŚÉņä£ Ļ┤Ćņ░░ļÉśņŚłļŗż. ļśÉĒĢ£ ņĢ╝Ē¢ēņä▒ņ£╝ļĪ£ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓Ģņ£╝ļĪ£ Ļ┤Ćņ░░ĒĢśĻĖ░ ņ¢┤ļĀĄļŗżĻ│Ā ņĢīļĀżņ¦ä Anguilla japonica (ļ▒Ćņןņ¢┤) [21]ļŖö 2Ļ░£ ĒĢśņ▓£ņŚÉņä£, ļśÉĒĢ£ ĻĄŁļé┤ ņłśĻ│äņŚÉņä£ ĒØöĒ׳ ņä£ņŗØĒĢśļŖö 1ĻĖēņłś ņ¦ĆĒæ£ ņ¢┤ņóģņØĖ Rhynchocypris oxycephalus (ļ▓äļōżņ╣ś), Misgurnus anguillicaudatus (ļ»ĖĻŠĖļ”¼), ļīĆĒśĢ ņ¢┤ņóģņØĖ Channa argus (Ļ░Ćļ¼╝ņ╣ś)ņÖĆ Silurus asotus (ļ®öĻĖ░)ļŖö 3Ļ░£ ĒĢśņ▓£ņŚÉņä£ eDNA ļČäņäØņŚÉņä£ļ¦ī Ļ▓ĆņČ£ļÉśņŚłļŗż.
ņāØļ¼╝ņĖĪņĀĢļ¦ØņŚÉ ļö░ļź┤ļ®┤ 3Ļ░£ ĒĢśņ▓£ ļ¬©ļæÉņŚÉņä£ Zacco platypus (Ēö╝ļØ╝ļ»Ė)Ļ░Ć ņĀÉņ£Āņ£© 39.7 ~ 52.4%ļĪ£ ņĀłļīĆņĀüņ£╝ļĪ£ ļ¦ÄņØ┤ Ļ┤Ćņ░░ļÉśņŚłĻ│Ā eDNA ļČäņäØņŚÉņä£ļÅä 10.3 ~ 21.2%ļĪ£ ļæÉ ļ░®ļ▓Ģ ļ¬©ļæÉņŚÉņä£ ņÜ░ņĀÉņóģņØś ĒĢśļéśļĪ£ ļéśĒāĆļé¼ļŗż. ņĢłņä▒ņ▓£ņŚÉņä£ļŖö Pseudoras-bora parva (ņ░ĖļČĢņ¢┤), Rhodeus uyekii (Ļ░üņŗ£ļČĢņ¢┤), Rhodeus ocellatus (ĒØ░ņżäļé®ņżäĻ░£)Ļ░Ć ņāüļīĆņĀüņ£╝ļĪ£ ļ¦ÄņØ┤ Ļ┤Ćņ░░ļÉśņŚłĻ│Ā eDNAņŚÉņä£ļŖö Acheilognathus rhombeus (ļé®ņ¦Ćļ”¼), Carassius auratus (ļČĢņ¢┤), Pseudorasbora parva (ņ░ĖļČĢņ¢┤)ņØś read ņłśĻ░Ć Ēö╝ļØ╝ļ»Ėļ│┤ļŗż ļ¦ÄņĢśļŗż. ļ│ĄĒĢśņ▓£ņŚÉņä£ļŖö Pseudogobio esocinus (ļ¬©ļלļ¼┤ņ¦Ć)Ļ░Ć Ēö╝ļØ╝ļ»ĖņØś ļÆżļź╝ ņØ┤ņŚłļŖöļŹ░ eDNAņŚÉņä£ļŖö Carassius auratus (ļČĢņ¢┤), Cyprinus carpio (ņ×ēņ¢┤), Acheilognathus rhombeus (ļé®ņ¦Ćļ”¼) ļō▒ņØ┤ ņāüļīĆņĀüņ£╝ļĪ£ ļ¦ÄņĢśļŗż. Ļ▓ĮņĢłņ▓£ņŚÉņä£ļŖö Ēö╝ļØ╝ļ»Ė ņØ┤ņÖĖņŚÉ Pungtungia herzi (ļÅīĻ│ĀĻĖ░), Rhinogobius sp. (ļ░Ćņ¢┤), Rhodeus uyekii (Ļ░üņŗ£ļČĢņ¢┤) Ļ░Ć ļ¦ÄņØ┤ Ļ┤Ćņ░░ļÉśņŚłņ£╝ļ®░ eDNAņŚÉņä£ļŖö Pseudorasbora parva (ņ░ĖļČĢņ¢┤), Rhinogobius sp. (ļ░Ćņ¢┤) ļō▒ņØ┤ ļ¦ÄņØ┤ ĒÖĢņØĖļÉśņŚłļŗż.
eDNA ļČäņäØņŚÉņä£ Ļ▓ĆņČ£ļÉ£ read ņłśņÖĆ ņāØļ¼╝ņĖĪņĀĢļ¦Ø ņ×ÉļŻīļź╝ ņØ┤ņÜ®ĒĢ┤ņä£ ĒÆŹļČĆļÅä (richness), ļŗżņ¢æņä▒ (Shannon diversity index) ņ¦Ćņłśļź╝ ņé░ņČ£ĒĢśņŚ¼ ļéśĒāĆļāłņ£╝ļ®░ ņāØļ¼╝ņĖĪņĀĢļ¦Ø ņ×ÉļŻīļŖö 3ļģä ĒÅēĻĘĀņØ┤ļŗż (Table 3). Ļ┤Ćņ░░ļÉ£ Ļ░£ņ▓┤ņłśļŖö 83~174ļĪ£ ņ░©ņØ┤Ļ░Ć Ēü¼ļéś eDNA ļ®öĒāĆļ░öņĮöļö®ņŚÉ ņØśĒĢ£ read ņłśļŖö 63,057~68,145ļĪ£ Ēü¼ņ¦Ć ņĢŖņĢśļŗż. eDNA ļČäņäØļ▓ĢņŚÉņä£ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉ ļ╣äĒĢ┤ ĒÆŹļČĆļÅäļŖö 2ļ░░ ņØ┤ņāü, ļŗżņ¢æņä▒ ņ¦ĆņłśļŖö 1.507~1.901 vs 2.357~2.509ļĪ£ ļŹö Ēü¼ļ®░ Ļ┤Ćņ░░ļÉ£ ņ┤Ø Ļ░£ņ▓┤ņłśĻ░Ć ņĀüņØĆ ļ│ĄĒĢśņ▓£ņŚÉņä£ ļæÉ ļ░®ļ▓ĢņØś ņ░©ņØ┤Ļ░Ć Ļ░Ćņן ņ╗Ėļŗż. ļŗżņ¢æņä▒ ņ¦ĆņłśļŖö ĒÆŹļČĆļÅäņÖĆ ĻĘĀļō▒ļÅäļź╝ Ļ▓░ĒĢ®ĒĢ£ ņ¦ĆņłśļĪ£ Ļ░ÆņØ┤ Ēü┤ņłśļĪØ ļŗżņ¢æņä▒ņØ┤ ņ╗żņ¦äļŗżļŖö Ļ▓āņØä ņØśļ»ĖĒĢśļ®░ ļæÉ ļ░®ļ▓ĢņØ┤ ņ░©ņØ┤Ļ░Ć ļéśļŖö ņØ┤ņ£ĀļŖö eDNA ļČäņäØļ▓ĢņØ┤ ļŹö ļ¦ÄņØĆ ņłśņØś ņ¢┤ļźś ņóģņØä Ļ▓ĆņČ£ĒĢśĻĖ░ ļĢīļ¼ĖņØ┤ļŗż. ĻĘĀļō▒ļÅä ņ¦ĆņłśļŖö ĻĄ░ņ¦æ ļé┤ ņóģ ĻĄ¼ņä▒ņØś ĻĘĀņØ╝ĒĢ£ ņĀĢļÅäļź╝ ļéśĒāĆļé┤ļŖö ņ¦ĆņłśņØ┤ļ®░ ļæÉ ļ░®ļ▓ĢņØä ĒåĄĒĢ┤ ņ¢╗ņØĆ Ļ▓░Ļ│╝ļŖö ņĢłņä▒ņ▓£(0.748 vs 0.761)ņŚÉņä£ļŖö ļ╣äņŖĘĒĢśļéś ļ│ĄĒĢśņ▓£(0.664 vs 0.752)Ļ│╝ Ļ▓ĮņĢłņ▓£(0.731 vs 0.694)ņŚÉņä£ļŖö ņ░©ņØ┤Ļ░Ć ņ╗Ėļŗż. eDNAņØś ļåŹļÅäĻ░Ć ņ¢┤ļźśņØś ļŗżņ¢æņä▒ ļ░Å ņāØņ▓┤ļ¤ēĻ│╝ ņ¢æņØś ņāüĻ┤ĆĻ┤ĆĻ│äĻ░Ć ņ׳ļŗżļŖö ļŗżņłśņØś ņŚ░ĻĄ¼Ļ░Ć ņ׳ņ£╝ļéś[22] ļČĆņĀĢņĀüņØĖ ņŚ░ĻĄ¼Ļ▓░Ļ│╝ ļśÉĒĢ£ ņĪ┤ņ×¼ĒĢ£ļŗż[23]. ņØ┤ļ▓ł ņĪ░ņé¼ņŚÉņä£ļÅä ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉ ļö░ļźĖ ņÜ░ņĀÉ ņ¢┤ņóģ (39.7~52.4%) Zacco platypus (Ēö╝ļØ╝ļ»Ė)Ļ░Ć ļ®öĒāĆļ░öņĮöļö® read ņłśļÅä ļ¦ÄņĢśņ£╝ļéś (ņĢłņä▒ņ▓£: 10.3%, ļ│ĄĒĢśņ▓£: 21.2%, Ļ▓ĮņĢłņ▓£: 19.3%), ņĢłņä▒ņ▓£ņŚÉņä£ļŖö Carassius auratus (ļČĢņ¢┤, 16.9%)ņÖĆ Pseudorasbora parva (ņ░ĖļČĢņ¢┤, 12.0%)ņØś read ņłśĻ░Ć ļŹö ļ¦ÄņĢśĻ│Ā, ļ│ĄĒĢśņ▓£ņŚÉņä£ļŖö Carassius auratus (ļČĢņ¢┤, 19.8%)ņÖĆ Cyprinus carpio (ņ×ēņ¢┤, 14.6%)Ļ░Ć ļ╣äĻĄÉņĀü ļ¦ÄņØ┤ Ļ▓ĆņČ£ļÉśņŚłĻ│Ā, Ļ▓ĮņĢłņ▓£ņŚÉņä£ļŖö Pseudorasbora parva (ņ░ĖļČĢņ¢┤, 21.6%)ņØś read ņłś ļ╣äņ£©ņØ┤ ļŹö ļåÆņĢśļŗż. ļö░ļØ╝ņä£ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉ ņØśĒĢ£ Ļ░£ņ▓┤ņłśņÖĆ ļ®öĒāĆļ░öņĮöļö®ņŚÉ ņØśĒĢ£ read ņłśņØś ņāüĻ┤ĆĻ┤ĆĻ│äļŖö ņ×ģņ”ØĒĢĀ ņłś ņŚåņŚłļŗż. ļŗżļ¦ī ņāüļīĆņĀüņØĖ ļ╣äĻĄÉ[24]ļź╝ ņ£äĒĢ┤ ļ░śņĀĢļ¤ēņĀü (semi-quantitative)ņ£╝ļĪ£ ņØ┤ņÜ®ĒĢĀ ņłś ņ׳ņ£╝ļ®░ read ņłśņØś ņāüļīĆņĀü ļ╣äņ£©ņØä ņØ┤ņÜ®ĒĢśņŚ¼ ĒÆŹļČĆĒĢ£ (abundant) ņóģĻ│╝ ĒؼĻĘĆĒĢ£ (rare) ņóģņØä ĻĄ¼ļČäĒĢśĻ▒░ļéś[5] ĒĢśņ▓£ ņāØĒā£Ļ│ä ļ│┤ņĀäņØä ņ£äĒĢ£ Ļ┤Ćļ”¼ļīĆņ▒ģ ņłśļ”ĮņŚÉļÅä ņ£ĀņÜ®ĒĢśĻ▓ī ņé¼ņÜ®ļÉĀ ņłś ņ׳ņØä Ļ▓āņØ┤ļŗż[25]. Ēśäņ×¼ ĻĄŁļé┤ ĒĢśņ▓£ņØś ņ¢┤ļźśņĪ░ņé¼ņŚÉņä£ļŖö ļīĆļČĆļČä Ēł¼ļ¦ØĻ│╝ ņĪ▒ļīĆĻ░Ć ĒÖ£ņÜ®ļÉśĻ│Ā ņ׳ņ£╝ļ®░ ņØ┤ļ¤¼ĒĢ£ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢļÅä ņ▒äņ¦æņØ┤ļéś ļÅÖņĀĢ Ļ│╝ņĀĢņŚÉņä£ ļ░£ņāØĒĢĀ ņłś ņ׳ļŖö ĒÄĖņ░© (bias)ļĪ£ ņØĖĒĢ£ ņĀĢļ¤ēņĀü ļČäņäØņŚÉļŖö ĒĢ£Ļ│äĻ░Ć ņ׳ļŗż[26]. ņŗżņĀ£ļĪ£ ņĢłņä▒ņ▓£, ļ│ĄĒĢśņ▓£, Ļ▓ĮņĢłņ▓£ņØś ņ¢┤ļźś ĻĄ░ņ¦æņĪ░ņé¼ņŚÉņä£ ņ▒äņ¦æļÉ£ Ļ░£ņ▓┤ņłśĻ░Ć 3ļģä ļÅÖņĢł 83~174ļĪ£ ļ¦Äņ¦Ć ņĢŖĻĖ░ ļĢīļ¼ĖņŚÉ Ļ░£ņ▓┤ņłśĻ░Ć ņĀüņØĆ ĒؼĻĘĆĒĢ£ ņóģņØś Ļ▓ĮņÜ░ņŚÉļŖö Ļ┤Ćņ░░ĒĢśĻĖ░ ņ¢┤ļĀżņÜĖ Ļ▓āņ£╝ļĪ£ ņśłņāüļÉśļ®░ ļÅÖņĀĢņŚÉņä£ļÅä ņśżļźśĻ░Ć ļ░£ņāØĒĢĀ Ļ░ĆļŖźņä▒ ļśÉĒĢ£ ņāüņĪ┤ĒĢ£ļŗż. ņØ┤ļ¤¼ĒĢ£ Ļ▓░Ļ│╝ļĪ£ ļ│╝ ļĢī eDNAļź╝ ņĀüņĀłĒĢśĻ▓ī ĒÖ£ņÜ®ĒĢ£ļŗżļ®┤ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢņŚÉņä£ ļåōņ╣śļŖö ņ¢┤ļźśļź╝ ĒśäņןņŚÉņä£ņØś ņ¦¦ņØĆ ņ▒äņłśņŗ£Ļ░ä ļ░Å ņĀüņØĆ ņŗ£ļŻīļ¤ēņ£╝ļĪ£ ļČäņäØņØ┤ Ļ░ĆļŖźĒĢśļ®░, 79% ņØ┤ņāü Ļ░ÖņØĆ ņ¢┤ņóģņØ┤ Ļ▓ĆņČ£ļÉśņ¢┤ņä£ ĒĢśņ▓£ņØś ņ¢┤ļźś ĻĄ░ņ¦æ ļČäņäØņŚÉ ņĀäĒåĄņĀüņØĖ ļ░®ļ▓ĢĻ│╝ ĒĢ©Ļ╗ś ĒÖ£ņÜ®ĒĢĀ ņłś ņ׳ņØä Ļ▓āņ£╝ļĪ£ ĒīÉļŗ©ļÉ£ļŗż.
ļ│Ė ņŚ░ĻĄ¼ņŚÉņä£ļŖö ĒĢśņ▓£ ņ¢┤ļźś ĻĄ░ņ¦æ ļČäņäØņØä ņ£äĒĢ┤ eDNAļź╝ ņØ┤ņÜ®ĒĢ£ ļČäņ×ÉņāØļ¼╝ĒĢÖņĀü ĻĖ░ļ▓ĢņØä ņĀüņÜ®ĒĢśņśĆļŗż. eDNA ļ®öĒāĆļ░öņĮöļö®ņØĆ ļ»╝ļ¼╝Ļ│ĀĻĖ░ ņāØĒā£ĒĢÖņŖĄĻ┤Ć ņŗ£ļŻīņŚÉ ļīĆĒĢ┤ 80%ņØś ņĀĢĒÖĢņä▒ņØä ļ│┤ņśĆļŗż. ļśÉĒĢ£ ĒĢśņ▓£ņØś ņ¢┤ļźś ņāØļ¼╝ņĖĪņĀĢļ¦ØņŚÉņä£ Ļ│╝Ļ▒░(2019 ~ 2021)ņŚÉ Ļ┤Ćņ░░ļÉśņŚłļŹś ņ¢┤ņóģņØś 79.2 ~ 87.5%ļź╝ Ļ▓ĆņČ£ĒĢśņŚ¼ ĒĢśņ▓£ ņ¢┤ļźś ĻĄ░ņ¦æ ļČäņäØņŚÉ eDNA ĒÖ£ņÜ® Ļ░ĆļŖźņä▒ņØä ļ│┤ņŚ¼ņŻ╝ņŚłļŗż. ĒĢ£Ļ│äņĀÉņ£╝ļĪ£ļŖö Mifish primerĻ░Ć ņ”ØĒÅŁĒĢśļŖö ņ£ĀņĀäņ×É ļČĆļČäņØ┤ 163 ~ 185bpļĪ£ ņ¦¦ņĢäņä£ Ļ░ÖņØĆ ņ£ĀņĀäņ×É ņŚ╝ĻĖ░ņä£ņŚ┤ņØä Ļ░¢ļŖöņóģņØ┤ļéś ņÜ░ļ”¼ļéśļØ╝ Ļ│Āņ£Āņóģ[11]ņØś Ļ▓ĮņÜ░ņŚÉ ĻĄ¼ļČäņØ┤ ņ¢┤ļĀĄĻĖ░ ļĢīļ¼ĖņŚÉ ĒŖ╣ņĀĢĒĢ£ ņóģļōżņØś ņĀĢĒÖĢĒĢ£ ļÅÖņĀĢņØä ņ£äĒĢ┤ņä£ļŖö ļ│┤ņÖäņĀüņØĖ primerņØś Ļ░£ļ░£ļÅä ĒĢäņÜöĒĢśļŗż. ņśłļź╝ ļōżņ¢┤ Mifish-tuna primerļŖö Thunnus (ļŗżļ×æņ¢┤ ņåŹ)ņŚÉ ņåŹĒĢśļŖö 7ņóģņØä Ļ▓ĆņČ£ĒĢśĻĖ░ ņ£äĒĢ┤ Ļ░£ļ░£ļÉśņŚłĻ│Ā[14], ļŗżļźĖ ņ£ĀņĀäņ×Éļź╝ ļīĆņāüņ£╝ļĪ£ ĒĢśļŖö primerļź╝ ņĪ░ĒĢ®ĒĢśņŚ¼ ņé¼ņÜ®ĒĢśļ®┤ ļŹö ļ¦ÄņØĆ ņ¢┤ņóģņØä Ļ▓ĆņČ£ĒĢĀ ņłś ņ׳ņŚłļŗż[27]. ļśÉĒĢ£ Ļ░Ćļ¼╝ņ╣ś, ļ®öĻĖ░ ļō▒ ĻĄŁļé┤ņŚÉ ņä£ņŗØĒĢśņ¦Ćļ¦ī ņĀäĒåĄņĀüņØĖ ņ▒äņ¦æ ļ░®ļ▓Ģņ£╝ļĪ£ ĒÖĢņØĖĒĢśņ¦Ć ļ¬╗Ē¢łļŹś ļīĆĒśĢ ņ¢┤ņóģļÅä ņØ┤ļ▓ł eDNA ļČäņäØņŚÉņä£ļŖö Ļ▓ĆņČ£ļÉśņŚłĻĖ░ ļĢīļ¼ĖņŚÉ ļæÉ ļ░®ļ▓ĢņØś ņāüĒśĖ ļ│┤ņÖä ĒÖ£ņÜ®ļÅä ĒĢäņÜöĒĢśļŗż. eDNA ļČäņäØņØĆ ĒśäņןņŚÉņä£ ĒŖ╣ļ│äĒĢ£ ņןļ╣ä ņŚåņØ┤ ņ¦¦ņØĆ ņŗ£Ļ░ä(~10ļČä)Ļ│╝ ņĀüņØĆ ņŗ£ļŻīļ¤ē (300 ~ 500 mL)ņ£╝ļĪ£ ņĀäĒåĄņĀüņØĖ ņ▒äņ¦æ ļ░®ļ▓Ģļ│┤ļŗż ļ¦ÄņØĆ ņóģļźśņØś ņ¢┤ļźś Ļ▓ĆņČ£ņØ┤ Ļ░ĆļŖźĒ¢łļŗż. ļśÉĒĢ£ Ēśäņן ņĪ░ņé¼ Ļ│╝ņĀĢņØś ņ£äĒŚśņä▒, ņĪ░ņé¼ Ļ│╝ņĀĢ ņżæņØś ļ®Ėņóģņ£äĻĖ░ ņ¢┤ļźś ĒÅÉņé¼, ņĪ░ņé¼ņ×ÉņØś ņłÖļĀ©ļÅäņŚÉ ļö░ļźĖ ņśżņ░©ļź╝ Ļ░Éņåīņŗ£Ēé¼ ņłś ņ׳ņ¢┤ņä£ ļ¬ć Ļ░Ćņ¦Ć ĒĢ£Ļ│äņĀÉņŚÉļÅä ļČłĻĄ¼ĒĢśĻ│Ā ļ│┤ņÖäņĀüņØ┤Ļ│Ā ļīĆņĢłņĀüņØĖ ņĀæĻĘ╝ ļ░®ņŗØņ£╝ļĪ£ ĒÖ£ņÜ®ļÉĀ ņłś ņ׳ņØä Ļ▓āņØ┤ļŗż. ļ│Ė ņŚ░ĻĄ¼ņŚÉņä£ ļČäņäØĒĢśĻĖ░ ņ¢┤ļĀżņøĀļŹś ņÜ░ļ”¼ļéśļØ╝ Ļ│Āņ£Āņóģ ļō▒ ĒŖ╣ņĀĢĒĢ£ ņ¢┤ļźś ņóģņŚÉ ļīĆĒĢ£ ņ£ĀņĀäņ×É ļŹ░ņØ┤Ēä░ļź╝ ņČĢņĀüĒĢśĻ│Ā ņĀüņĀłĒĢ£ ņŗżĒŚśļ▓ĢņØä ņĀüņÜ®ĒĢ£ļŗżļ®┤, ņĀüņØĆ ļģĖļĀźņ£╝ļĪ£ ņןĻĖ░ņĀüņ£╝ļĪ£ ļŹö ņĀĢĒÖĢĒĢ£ ņāØļ¼╝ļŗżņ¢æņä▒ ņ×ÉļŻīļź╝ ĒÖĢļ│┤ĒĢĀ ņłś ņ׳ņØä Ļ▓āņ£╝ļĪ£ ĻĖ░ļīĆļÉ£ļŗż.
Ļ░Éņé¼ņØś ĻĖĆ
ļ│Ė ļģ╝ļ¼ĖņØĆ ĒÖśĻ▓ĮļČĆņØś ņ×¼ņøÉņ£╝ļĪ£ ĻĄŁļ”ĮĒÖśĻ▓ĮĻ│╝ĒĢÖņøÉņØś ņ¦ĆņøÉņØä ļ░øņĢä ņłśĒ¢ēĒĢśņśĆņŖĄļŗłļŗż (NIER-2022-01-03-014).
Table┬Ā1.
Detection of freshwater fish species in aquarium using eDNA metabarcoding
Table┬Ā2.
Comparison of fish species detection in three rivers using traditional method (T, 2019~2021) and eDNA metabarcoding (D)
Table┬Ā3.
Comparison of fish diversity index between traditional method (T) and eDNA metabarcoding (D)
ņ░ĖĻ│Āļ¼ĖĒŚī
1. P. Taberlet, E. Coissac, M. Hajibabaei, and H. Rieseberg, ŌĆ£Environmental DNAŌĆØ, Molecular Ecology, 2012, 21, 1789-1793.


2. P. Taberlet, and E. Coissac, ŌĆ£Environmental DNA - An emerging tool in conservation for monitoring past and present biodiversityŌĆØ, Biological Conservation, 2015, 183, 4-18.

3. M. A. Barnes, C. R. Turner, C. L. Jerde, M. A. Renshaw, W. L. Chadderton, and D. M. Lodge, ŌĆ£Environmental conditions influence eDNA persistence in aquatic systemsŌĆØ, Environmental Science & Technology, 2014, 48, 1819-1827.


4. K. Deiner, E. A. Fronhofer, E. M├żchler, J.-C. Walser, and F. Altermatt, ŌĆ£Environmental DNA reveals that rivers are conveyer belts of biodiversity informationŌĆØ, Nature Communication, 2016, 7, 12544.




5. A. Valentini, P. Taberlet, C. Miaud, R. Civade, J. Herder, P. F. Thomsen, E. Bellemain, A. Besnard, E. Coissac, F. Boyer, C. Gaboriaud, P. Jean, N. Poulet, N. Roset, G. H. Copp, P. Geniez, D. Pont, C. Argillier, J.-M. Baudoin, T. Peroux, A. J. Crivelli, A. Olivier, M. Acqueberge, M. L. Brun, P. R. M├Ėller, E. Willerslev, and T. Dejean, ŌĆ£Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcodingŌĆØ, Molecular Ecology, 2016, 25, 929-942.


6. E. E. Sigsgaard, I. B. Nielsen, H. Carl, M. A. Krag, S. W. Knudsen, Y. Xing, T. H. Holm-Hansen, P. R. M├Ėller, and P. F. Thomsen, ŌĆ£Seawater environmental DNA reflects seasonality of a coastal fish communityŌĆØ, Marine Biology, 2017, 164, 128.


7. E.-B. Kim, H. Sagong, J.-H. Lee, G. Kim, D.-H. Kwon, Y. Kim, and H.-W. Kim, ŌĆ£Environmental DNA Metabarcoding Analysis of Fish Assemblages and Phytoplankton Communities in a Furrowed Seabed Area Caused by Aggregate MiningŌĆØ, Frontiers in Marine Science, 2022, 9.

8. A. R. Kim, T.-H. Yoon, C. I. Lee, C.-K. Kang, and H.-W. Kim, ŌĆ£Metabarcoding Analysis of Ichthyoplankton in the East/Japan Sea Using the Novel Fish-Specific Universal Primer SetŌĆØ, Frontiers in Marine Science, 2021, 8.

9. G. Kim, and Y. Song, ŌĆ£Identification of Freshwater Fish Species in Korea Using Environmental DNA Technique - From the Experiment at the Freshwater Fish Ecological Learning Center in Yangpyeong, Gyeonggi DoŌĆØ, Journal of Environmental Impact Assessment, 2021, 30, 1-12.
10. Y.-K. Song, J.-H. Kim, S.-Y. Won, and C. Park, ŌĆ£Possibility in identifying species composition of fish communities using the environmental DNA metabarcoding technique - with the preliminary results at urban ecological streamsŌĆØ, Journal of the Korean Society of Environmental Restoration Technology, 2019, 22, 125-138.
11. J.-H. Kim, H. Jo, M.-H. Chang, S.-H. Woo, Y. Cho, and J.-D. Yoon, ŌĆ£Application of Environmental DNA for Monitoring of Freshwater Fish in KoreaŌĆØ, Korean Journal of Ecology and Environment, 2020, 53, 63-72.

12. M. J. Alam, N.-K. Kim, S. Andriyono, H.-K. Choi, J.-H. Lee, and H.-W. Kim, ŌĆ£Assessment of fish biodiversity in four Korean rivers using environmental DNA metabarcodingŌĆØ, PeerJ, 2020, 8, e9508.




13. M. Miya, Y. Sato, T. Fukunaga, T. Sado, J. Y. Poulsen, K. Sato, T. Minamoto, S. Yamamoto, H. Yamanaka, H. Araki, M. Kondoh, and W. Iwasaki, ŌĆ£MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine speciesŌĆØ, Royal Society Open Science, 2015, 2, 150088.




14. M. Miya, R. O. Gotoh, and T. Sado, ŌĆ£MiFish metabarcoding: a high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samplesŌĆØ, Fisheries Science, 2020, 86, 939-970.


15. Y. Sato, M. Miya, T. Fukunaga, T. Sato, and W. Iwasaka, ŌĆ£MitoFish and MiFish Pipeline: A Mitochondrial Genome Database of Fish with an Analysis Pipeline for Environmental DNA MetabarcodingŌĆØ, Molecular Biology and Evolution, 2018, 35 (6), 1553-1555.

16. K.-S. Kim, K.-Y. Kim, and J.-D. Yoon, ŌĆ£Efficiency Comparison of Environmental DNA Metabarcoding of Freshwater Fishes according to Filters, Extraction Kits, Primer Sets and PCR MethodsŌĆØ, Korean Journal of Ecology and Environment, 2021, 54, 199-208.

17. R. Hinlo, D. Gleeson, M. Lintermans, and E. Furlan, ŌĆ£Methods to maximise recovery of environmental DNA from water samplesŌĆØ, PloS one, 2017, 12, e0179251.



18. S. Tsuji, T. Takahara, H. Doi, N. Shibata, and H. Yamanaka, ŌĆ£The detection of aquatic macroorganisms using environmental DNA analysisŌĆöA review of methods for collection, extraction, and detectionŌĆØ, Environmental DNA, 2019, 1, 99-108.


19. L. Shu, A. Ludwig, and Z. Peng, ŌĆ£Standards for Methods Utilizing Environmental DNA for Detection of Fish SpeciesŌĆØ, Genes, 2020, 11, 296.



20. H.-T. Ruan, R.-L. Wang, H.-T. Li, L. Liu, T.-X. Kuang, M. Li, and K.-S. Zou, ŌĆ£Effects of sampling strategies and DNA extraction methods on eDNA metabarcoding: A case study of estuarine fish diversity monitoringŌĆØ, Zoological Research, 2022, 43, 92-204.
21. H. Itakura, R. Wakiya, S. Yamamoto, K. Kaifu, T. Sato, and T. Minamoto, ŌĆ£Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scaleŌĆØ, Aquatic Conservation: Marine and Freshwater Ecosystems, 2019, 29, 361-373.


22. M. L. Rourke, A. M. Fowler, J. M. Hughes, M. K. Broadhurst, J. D. DiBattista, S. Fielder, J. W. Walburn, and E. M. Furlan, ŌĆ£Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveysŌĆØ, Environmental DNA, 2022, 4, 9-33.


23. M. L. Rourke, J. W. Walburn, M. K. Broadhurst, A. M. Fowler, J. M. Hughes, D. S. Fielder, J. D. DiBattista, and E. M. Furlan, ŌĆ£Poor utility of environmental DNA for estimating the biomass of a threatened freshwater teleost; but clear direction for future candidate assessmentsŌĆØ, Fisheries Research, 2023, 258, 106545.

24. R. P. Kelly, J. A. Port, K. M. Yamahara, and L. B. Crowder, ŌĆ£Using Environmental DNA to Census Marine Fishes in a Large MesocosmŌĆØ, PloS one, 2014, 9, e86175.



25. T. Takahara, T. Minamoto, H. Yamanaka, H. Doi, and Z. Kawabata, ŌĆ£Estimation of Fish Biomass Using Environmental DNAŌĆØ, PloS one, 2012, 7, e35868.



- TOOLS
-
METRICS

-
- 1 Crossref
- 851 View
- 8 Download
-
Related articles in
J Environ Anal Health Toxicol -
Quality Control for the Analysis of Heavy Metals in Herbal Medicine2012 December;15(4)
Comparison of Pretreatment Methods for the Analysis of PCBs in Transformer Oil2010 September;13(3)